Abstract
Ever used a pair of molecular scissors? Restriction enzymes are molecular scissors that cut DNA into pieces. Find out which enzymes will cut, and where by making a restriction map. Then you can figure out what will happen if you change the sequence of the DNA. Will the same enzymes still cut the new DNA sequence?Summary

Objective
In this experiment you will determine if cutting a piece of DNA with a restriction enzyme depends upon the sequence of the DNA.Introduction
All living things come with a set of instructions stored in their DNA, short for deoxyribonucleic acid. Whether you are a human, rat, tomato, or bacteria, each cell will have DNA inside of it. DNA is the blueprint for everything that happens inside the cell of an organism, and each cell has an entire copy of the same set of instructions. The entire set of instructions is called the genome and the information is stored in a code of nucleotides (A, T, C, and G) called bases. Here is an example of a DNA sequence that is 12 base pairs long:
Notice that this piece of DNA has two sequences: one on the top and one on the bottom. DNA is double stranded, which means that it has two strands. The nucleotides of each of these strands are paired together in a particular way to match the other strand: A pairs with T and C pairs with G. If a nucleotide is paired according to these rules, it is called a match. But if the nucleotide is not paired properly, then it is called a mismatch.
The information stored in the DNA is coded into sets of nucleotide sequences called genes. Each gene is a set of instructions for making a specific protein. The protein has a certain job to do, called a function. Since different cells in your body have different jobs to do, many of the genes will be turned on in some cells, but not others. For example, some genes code for proteins specific to your blood cells, like hemoglobin. Other genes code for proteins specific to your pancreas, like insulin. Even though different genes are turned on in different cells, your cells and organs all work together in a coordinated way so that your body can function properly.
What if there is something wrong with one of your genes? This can cause problems for your body and how it functions. For example, people who have type I diabetes have problems making insulin. To help people with diabetes, scientists figured out a way to make insulin that diabetics can inject into their body. The insulin is made by a bacteria that has the human gene for insulin.
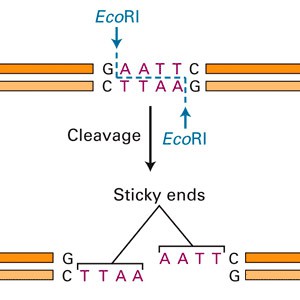
For scientists to study a gene, they need to be able to isolate it. The simplest way to isolate a gene is to cut it out and clone the gene into a small piece of bacterial DNA called a plasmid. How do you cut out a piece of DNA? A restriction enzyme is a protein that acts like a pair of molecular scissors to cut a DNA strand. The enzyme recognizes a certain DNA sequence where it will cut the DNA apart. Here is an example of a restriction enzyme called EcoRI that cuts DNA at a particular sequence, creating sticky ends:
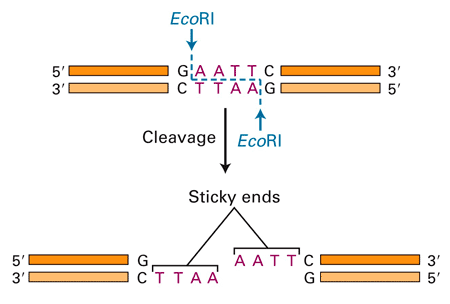
(Image from Biotechnology Online, 2007)
Once the DNA is cut apart, it can be put back together in different ways. For example, the gene for human insulin can be cut out and then recombined with the DNA of a bacteria. Then the bacteria can grow and make human insulin for people who need it to manage their diabetes. Cutting DNA apart using restriction enzymes is a very important step in the discovery and manufacturing of genes that become important pharmaceuticals, like the insulin gene.
In this experiment, you will investigate how restriction enzymes recognize different DNA sequences. You will use a computer program to generate random pieces of DNA sequence. Then you will use another program to test your sequence and look for restriction enzymes that will cut it (often called cutters). By comparing which restriction enzymes cut each unique DNA sequence, you can determine if changing the DNA sequence will change the restriction enzymes that cut it. If the DNA sequence does not affect the restriction enzyme that cuts it, then the different DNA sequences will have a lot of cutters in common. If the DNA sequence does affect the restriction enzyme that cuts it, then the different DNA sequences will have more unique cutters than common cutters.
Terms and Concepts
To do this type of experiment you should know what the following terms mean. Have an adult help you search the Internet, or take you to your local library to find out more!
- Biotechnology
- DNA
- Nucleotide (A, T, C, G)
- Sequence
- Strand
- Genome
- Gene
- Restriction enzyme
Questions
- What does a DNA sequence look like?
- How can I find out which restriction enzymes will cut a piece of DNA?
- Which restriction enzymes cut the DNA?
- If I change the DNA sequence, will the restriction enzymes also change?
Bibliography
This book is a good source of background information about the biotechnology behind this project:
- Brown, J. Kirk. Biotechnology: A Laboratory Skills Course. Student Edition. Hercules: Bio-Rad Laboratories, Inc, 2011.
- Learn how to cut and paste genes from Biotechnology Online:
- Biotechnology Online, 2007. Cutting and Pasting Genes, Biotechnology Australia, An Australian Government Initiative. Retrieved March 6, 2007.
- Learn all about the science of biotechnology from the Biotechnology Learning Center at the Children's Museum of Indianapolis. Be sure to watch the Buzzz About Biotechnology animation!
- BLC, 2007. "Biotechnology Learning Center," The Children's Museum of Indianapolis. Retrieved March 6, 2007.
- You will use this site to make short, randomized sequences of DNA:
Maduro, M., date unknown. Random DNA Sequence Generator, Department of Biology, University of California, Riverside. Retrieved March 6, 2007. - You will use this site to check your DNA to see which enzymes cut it and where:
NEB, 2007. NEBcutter, New England Biolabs (NEB). Retrieved March 6, 2007. - Here are two sites with some background information on restriction enzymes:
- Kimball, J., 2003. Restriction Enzymes, Andover, MA: Kimball's Biology Pages. Retrieved March 6, 2007.
- Wikipedia contributors, Restriction enzyme, Wikipedia, The Free Encyclopedia. Retrieved June 28, 2010.
Materials and Equipment
- A computer with internet connection
- Java-based web browser
- Lab notebook and pencil
- Printer
- Scissors
- Glue
Experimental Procedure
- The first step is to make a piece of DNA using the Random DNA Sequence Generator.
- Enter "20" in the box for the Size of DNA in base pairs (bp), and leave the setting for the GC content at 0.50 (which will give you half G+C and half A+T).
- Click the generate button and you will get a random piece of DNA shown in the text box:
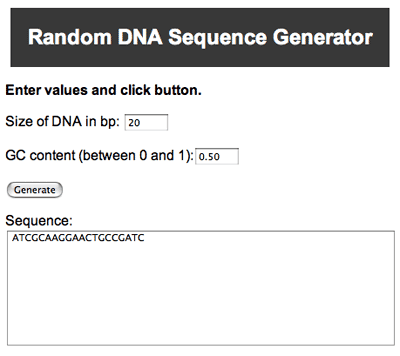
A random DNA sequence generator hosted on ucr.edu. This generator allows you to set variables such as the number of base pairs and how often G and C will show up in the end result.
- Double click in the text box to select your DNA sequence, then copy it to the clipboard by selecting "Edit" and then "Copy" from your file menu.
- The next step is to check your piece of randomly generated DNA to see which restriction enzymes will cut it using the NEBcutter program from New England Biolabs, a company that happens to sell (you guessed it) restriction enzymes!
- Click inside the text box and paste your DNA sequence from the clipboard by selecting "Edit" and "Paste" from the file menu. Leave all of the other settings to the default settings.
- Click the "Submit" button and you will get a page showing your piece of DNA and one or more enzymes that cut it. This is called a restriction enzyme map. Each arrow is a place where the enzyme will cut the DNA. Each restriction enzyme has a name that is written in code, for example EcoRI. You can click on a name to get more information about that enzyme. Also, notice that there is an arrow on both the top and bottom strand of DNA:
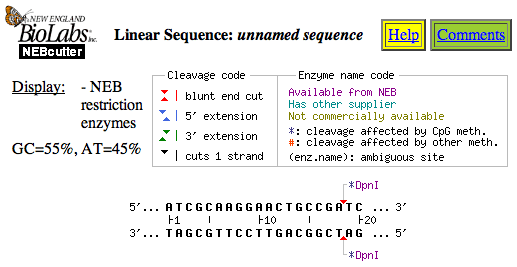
Neb.com hosts a NEBcutter program that allows users to upload a DNA sequence and it will return restriction enzymes that will be effective in cutting the DNA sequence.
- Print this page, cut it out, and paste it into your lab notebook for your records. Make an alphabetical list of the enzyme(s) that cut the DNA.
- Now you are ready to repeat steps 1-8 with a new DNA sequence. Just go back to the Random DNA Sequence Generator and start over.
- Repeat this experiment at least five different times. Each time you will make one new piece of DNA and test it for cutters by making a restriction enzyme map.
- Compare all of your sequences and the restriction enzyme maps, then ask yourself some questions:
- Is there an enzyme which shows up in more than one map?
- Is there any enzyme which shows up on all of the maps, that your DNA sequences have in common?
- Is there as sequence that only cuts one of the DNA sequences, that is unique?
- Draw a circle around any restriction enzymes which are common in all of the DNA sequences you tested. Count them and write the number in your lab notebook.
- Draw a square around any restriction enzyme which is unique to one of the DNA sequences you tested. Count them and write the number in your lab notebook.
- In this experiment, you have collected a lot of data in your notebook. Now you need to analyze your data and put it together into a story. The first step is to make a summary table of your experiment. Here is a sample summary table that you might want to use for this experiment:
| Name of Sequence | How Many Restriction Enzymes Cut? | How Many are Unique? | How Many are Common? |
|---|---|---|---|
| DNA#1 | |||
| DNA#2 | |||
| DNA#3 | |||
| DNA#4 | |||
| DNA#5 |
- Now you can make some graphs to show your data. For this experiment, you can make a bar graph to show the number of unique and common restriction enzymes for each DNA sequence you tried.
- Now make your conclusion by relating your results to your objective. Are there more unique cutters, or more cutters in common for the different DNA sequences? Did changing the DNA sequence change the restriction enzymes that cut the DNA? Do you think that restriction enzymes are unique to specific DNA sequences?
Ask an Expert
Global Connections
The United Nations Sustainable Development Goals (UNSDGs) are a blueprint to achieve a better and more sustainable future for all.
Variations
- Instead of generating a brand new random sequence each time, you can also make changes to the DNA sequence yourself. You can make subtle changes, or dramatic changes. You can try to add or change restriction enzyme sites. How will these kinds of changes affect your restriction map?
- You can test whether the length of the DNA will change the number of restriction enzymes that cut it. Just change the size of DNA in bp in step one from 20 bp to test a series of DNA sizes. Try 20 bp, 40 bp, 60 bp, and 100 bp. Which has more cutters, long sequences or short sequences?
- Some enzymes cut in the same place as other enzymes. By looking at the sequence of the cut site, can you explain why? Try comparing the cut site of restriction enzymes that cut in the same places as other restriction enzymes. What do you see?
- Sometimes, two restriction enzymes leave ends that can stick together. These are called sticky ends, and will match up to another piece of DNA with matching ends. Can you figure out how to use this experiment to generate different pieces of DNA and then put them together?
Careers
If you like this project, you might enjoy exploring these related careers:
Related Links
- Science Fair Project Guide
- Other Ideas Like This
- Biotechnology Project Ideas
- Genetic Engineering Project Ideas
- My Favorites