Abstract
Scientists recently found that some small drugs can stop infection by the deadly Ebola virus in its tracks. Lab researchers found that these drugs bind to a protein that the Ebola virus uses to enter our cells, and this is how infection is prevented. However, this also means that the bound protein no longer functions in our cells. How might these drugs accidentally disrupt important biological processes in our bodies? What other proteins might these drugs bind to? In this science project, you will explore how drugs that may someday be used to treat deadly diseases are tested to make sure that they do not unintentionally damage our bodies.Summary
Teisha Rowland, PhD, Science Buddies
Svenja Lohner, PhD, Science Buddies

Objective
Determine how untested drugs may affect important biological processes that they were not intended to.
Introduction
Recently, a breakthrough was made in our understanding of how the Ebola virus infects people. Many outbreaks of the Ebola virus have occurred in Africa, and infection is often deadly. You can see what the virus looks like in Figure 1. In August 2011, two different groups of researchers reported that in order to enter our cells and infect our bodies, the Ebola virus must bind to a protein called Niemann-Pick C1 ("NPC1" for short). To figure this out, one of the research groups (Carette et al.) first took a large number of cells and randomly disrupted different genes in the different cells. Eventually, the number of genes disrupted, or mutated, in all of the different cells combined was in the millions. The researchers exposed these mutated cells to the Ebola virus and then checked to see if any cells were resistant to infection. They analyzed the resistant cells to see what genes were mutated in these specific cells. They realized that the genes, especially NPC1, that were mutated in the resistant cells probably play key roles in infection. This is how the researchers found that NPC1 is essential for the Ebola virus to infect cells.
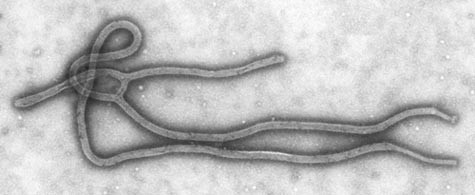 Image Credit: Wikimedia Commons / Public domain
Image Credit: Wikimedia Commons / Public domain
Figure 1. This is a picture of the deadly Ebola virus, taken with a very high magnification microscope (a transmission electron microscope). Recent research found that it can infect our cells using a protein called Niemann-Pick C1, which has become the target of anti-viral drugs.
NPC1 is found along the membrane of endosomes, which are small compartments in our cells that transport molecules from the outside of the cell to the inside. Inside the cell, molecules in endosomes are carried to lysosomes, which are compartments that break down molecules and cell debris. Normally, NPC1 is important for transporting cholesterol in cells, but the Ebola virus uses NPC1 to gain entry into the endosomes and causes the endosomes to burst, releasing the virus into the cell.
One of the research groups (Cóté et al) already developed two anti-viral drugs that can block infection by binding the NPC1 protein. To find these successful drugs, the researchers first tested a large number of small molecules on cells exposed to the Ebola virus to see if any of the small molecules could prevent infection. Sure enough, one small molecule, labeled 3.0, stopped infection. The researchers made 50 small molecules similar to this one and found that one of these 50, labeled 3.47, worked even better at preventing Ebola infection. (3.0 is technically a benzylpiperazine adamantine diamide molecule, and 3.47 is just like this molecule but has a methoxycarbonyl benzyl group added to it.)
While the researchers (Cóté et al) found that their drugs bind NPC1, and that this can block Ebola infection, extensive pharmaceutical testing still needs to be done before doctors can use these drugs to fight Ebola infection in people. For example, it needs to be determined whether the drugs bind other proteins that are similar to NPC1. Additionally, because researchers had the goal to study how the drugs prevent infection, using only cells grown in a lab, they did not find out how the drugs affect the overall health and function of the cells. For example, the drugs might interfere with important signaling pathways (biochemical pathways). Or the drugs may even affect the body of an animal as a whole. In summary, currently researchers do not know whether these will be good clinical drugs. In this science project, using bioinformatics tools (computer tools used to explore biological processes), you will explore how these Ebola virus drugs could bind non-target proteins, that is, proteins other than NPC1, and how disrupting the normal function of NPC1 and these non-target proteins could interfere with normal cellular and bodily functions. The questions that you will tackle here are exactly the ones researchers will be addressing, with the only difference being that researchers will be able to do both the bioinformatics work, what is covered in this science project, and testing of the resulting hypothesis in the lab.
Terms and Concepts
- Ebola virus
- Protein
- Genes
- Endosomes
- Lysosomes
- Drugs
- Small molecules
- Pharmaceutical
- Signaling pathways
- Bioinformatics
- Microarray
- Expression of genes
Questions
- How did the researchers determine that NPC1 is necessary for the Ebola virus to infect cells?
- What does NPC1 normally do in the cell?
- How does the Ebola virus take advantage of the normal function of NPC1 to gain entry into the cell?
- How specific do you think newly developed drugs are? Do you think they could bind many non-target proteins?
- Knowing what you do about the normal function of NPC1, what important cellular processes do you think the drugs might disrupt?
- Why is it important to test new drugs in animals before using them in humans?
Bibliography
To do this science project you will need to use these databases:
- Kyoto Encyclopedia of Genes and Genomes (KEGG). (n.d.). KEGG Pathway Database. Retrieved September 9, 2011.
- NCBI. (n.d.). BLAST: Basic Local Alignment Search Tool. Retrieved September 9, 2011.
- NCBI. (n.d.). Gene. Retrieved September 9, 2011.
These resources are a good place to start gathering information about drugs, the Ebola virus, and NPC1:
- Centers for Disease Control and Prevention. (n.d.). Ebola Hemorrhagic Fever. Retrieved September 9, 2011.
- Genetics Home Reference (n.d.). NPC1. Retrieved September 9, 2011.
- The Naked Scientists. (2011, August 24). Ebola virus blocked. Retrieved September 7, 2011.
- Sullivan, N., Yang, Z., and Nabel, G. (2003, September). Ebola Virus Pathogenesis: Implications for Vaccines and Therapies. Journal of Virology. Vol 77, No 18, 9733-9737. Retrieved September 9, 2011.
These are the two studies that discovered that the Ebola virus enters cells using the NPC1 protein and developed small molecule drugs to bind NPC1 and block infection:
- Carette, J. et al. (2011, August 24). Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. Published online. Retrieved September 7, 2011.
- Cóté, M. et al. (2011, August 24). Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. Published online. Retrieved September 7, 2011.
Materials and Equipment
- Computer with an Internet connection
- Lab notebook
Experimental Procedure
Identifying Non-Target Proteins
How accurately does a drug bind to its target protein (the protein it is supposed to bind to)? Are there other proteins the drug might bind to that it should not? In this part of the science project you will be looking at what non-target proteins the Ebola drugs might interact with, based on how similar the target protein is to other proteins.
- Learn as much as you can about the Niemann-Pick CI gene and protein. You can do this using the
NCBI Gene Database.
- Go to the NCBI Gene database website. Use the Niemann-Pick C1 gene name, NPC1, for your search. Write "NPC1 homo sapiens" into the search box, as you want to search for the NPC1 gene in humans. Then click "Search."
- The resulting page may have a long list of related results. The top results are usually the most relevant ones. Make sure to choose the top result that says "NPC1" and is in humans (Homo sapiens). Click on the gene link to open the gene page.
- What does the gene page tell you about the NPC1 gene and protein? What disease do defects in this gene cause? What part(s) of the cell is the NPC1 protein located in?
- Hint: You may also want to search through other databases, such as by using the
Genetics Home Reference tool, to find out more about NPC1
and the disease associated with it.
- In the "Genes" section, you can look up specific genes and find a variety of information on them, including their effects on health and genetics. Click on the letter "N" and scroll down until you see "NPC1" and click on it. Read the information about the NPC1 gene to find out more about what it does and what diseases it is linked to.
- Retrieve the amino acid sequence of the NPC1 protein.
- From within the NCBI Gene page about NPC1, navigate to the "Related Information" sidebar on the right-hand side of the page. Click on the "Protein" link.
- Click on the top result for the full NPC1 protein, as shown in Figure 2, circled in green. Make sure to select the full protein and not a partial sequence.
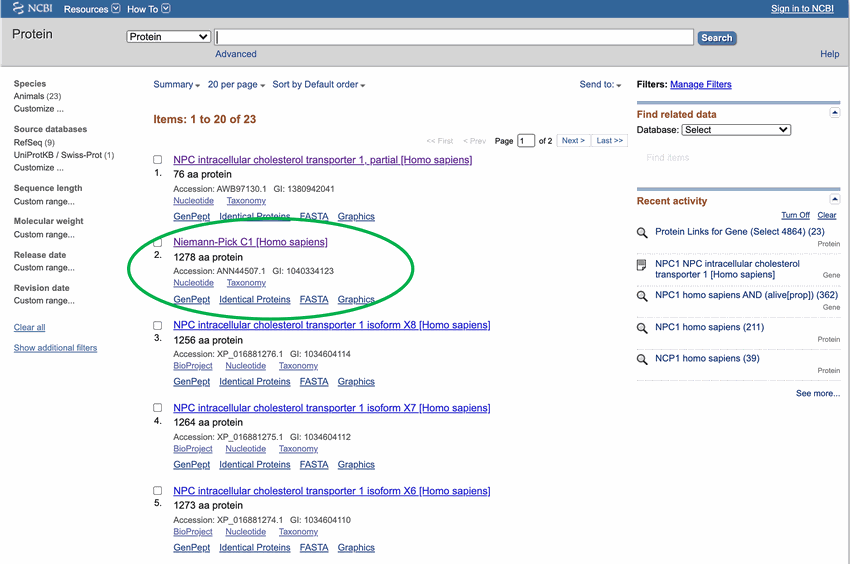 Image Credit: National Center for Biotechnology Information (NCBI)
Image Credit: National Center for Biotechnology Information (NCBI)The search results on the webpage ncbi.nlm.nih.gov returns a list of proteins and variations of those proteins. The Niemann-Pick C1 protein has variations that may cause some to click on the wrong link. Each search result has a unique number under the link so the correct link can be identified. The number for this link is GI: 255652944.
Figure 2. Searching for the amino acid sequence for the Niemann-Pick C1 protein will generate many results. Many of these are variants of the NPC1 protein. Pick the top result that is in humans ([Homo sapiens]), circled in green.
- On the protein page, scroll down to the bottom, where the amino acid sequence for the protein is given. Copy the entire amino acid sequence (including the numbers at the start of each line), as shown in Figure 3, circled in green. It is a good idea to save this sequence in a word or text file in case you need it later.
 Image Credit: Teisha Rowland, Science Buddies / Science Buddies
Image Credit: Teisha Rowland, Science Buddies / Science BuddiesNear the bottom of the NPC1 protein page on the NCBI webpage there is an amino acid sequence listed.
Figure 3. The amino acid sequence of the protein will be located at the bottom of the protein page for the NPC1 protein. Scroll down to the bottom and select and copy the amino acid sequence, circled in green, so that you can compare this sequence to other protein sequences in the following steps of the Experimental Procedure.
- Next, use the NCBI Basic Local Alignment Search Tool (BLAST) to identify any other human proteins that have a similar amino acid sequence. If there are any, they are candidates for proteins that the Ebola drugs might also bind to and interfere with.
- You will find that there are several versions of BLAST. Since you want to use a protein sequence to search a protein database, click on "Protein BLAST" under the "Web BLAST" heading.
- Fill out the protein BLAST query form. Once filled out, it should look like the one in Figure 4.
- Copy and paste the NPC1 protein sequence into the box labeled "Enter accession number, gi, or FASTA sequence".
- In the "Job title" box, type "NPC1 protein."
- In the "Database" box, choose "Reference proteins (refseq_protein)" from the drop-down menu.
- In the "Organism" box, write "homo sapiens" and select "Homo sapiens (taxid:9606)" from the list. This will restrict your search to proteins in humans only.
- Under "Algorithm," click on "blastp" (which is protein vs. protein).
- Keep all the other search parameters including the Algorithm parameters at their default settings.
- Next to the BLAST button at the bottom of the page, check the box "show results in a new window."
- Click on BLAST to start the search.
 Image Credit: National Center for Biotechnology Information (NCBI)
Image Credit: National Center for Biotechnology Information (NCBI)In the blastp tab in the BLAST tool there is a large textbox at the top of left of the page where amino acid sequences can be entered to search for related sequences. In the center of the page a drop down menu allows you to search through different databases.
Figure 4. Using the NCBI BLAST tool, you can search for proteins with similar amino acids by entering the amino acid sequence you want to search for.
- Be patient. It will take a minute or two for the BLAST search to finish. The results page will resemble the one in Figure 5. This page gives you a lot of information on how the amino acid sequence you submitted matches other sequences in the database.
- On the top left of the BLAST results page you will find the summary section (blue in Figure 5), which provides information on different aspects of your search. On the top right there is a box that allows you to filter your results based on certain criteria (red in Figure 5). Below the top section, the BLAST results are shown (yellow in Figure 5). There are four different tabs called "Description," Graphic Summary," "Alignments," and "Taxonomy." Each tab presents the search results in a different way.
- The "Description" tab contains a summary table of hits found by BLAST and is the default tab shown.
- The "Graphic Summary" tab shows a color key of the alignments. The color key shows the degree of similarity for the sequences.
- The "Alignment" section contains the detailed pairwise alignments between query and database sequences.
- The "Taxonomy" section provides details of the taxonomic distribution of matches BLAST found.
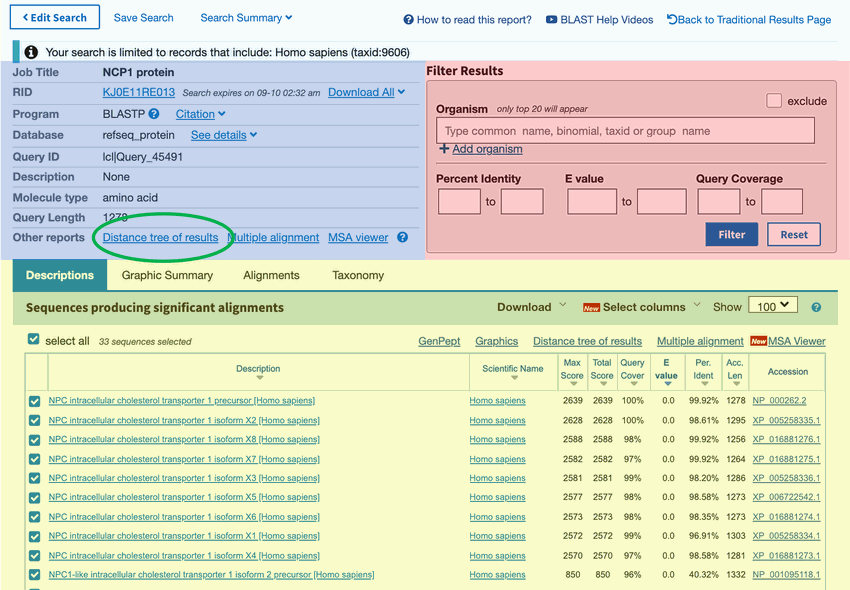 Image Credit: National Center for Biotechnology Information (NCBI)
Image Credit: National Center for Biotechnology Information (NCBI)The results page in the BLAST tool on the NCBI webpage shows a list of protein sequences that match a search term. On the page there are links to additional reports such as a search summary, taxonomy report, distance tree of results, and multiple alignments.
Figure 5. Searching for similar amino acid sequences on the NCBI BLAST website will probably generate many results that match your sequence to varying degrees. Clicking on the "Distance tree of results" link, circled in green, will take you to a helpful visual representation of your results.
- At the top of this results page, in the blue section, click on the "Distance tree of results" link, as shown in Figure 5, circled in green.
- On the distance tree of results page, hover over each green arrow at the end of the tree branches, and click on the "Expand/Collapse" link until you completely expand all of the branches, as shown in Figure 6.
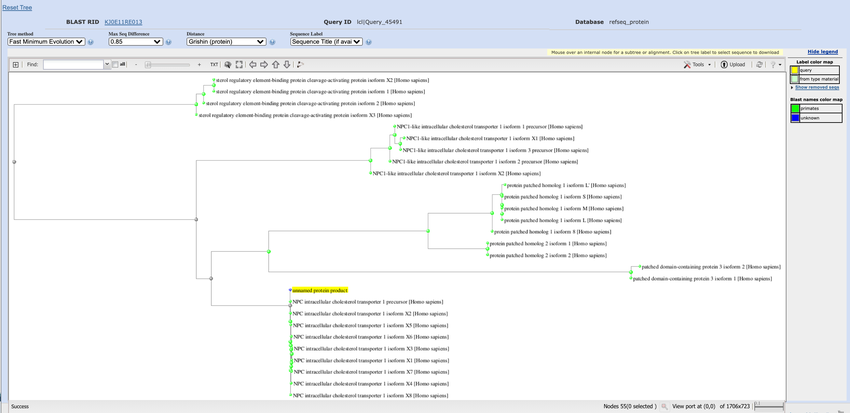 Image Credit: National Center for Biotechnology Information (NCBI)
Image Credit: National Center for Biotechnology Information (NCBI)The BLAST tool on the NCBI webpage can generate distance trees that map relationship between proteins based on shared sequences. Clicking on an arrow at the end of a branch allows results to be expanded and will list the names of proteins related to a specific amino acid.
Figure 6. By expanding the branches in the "Distance tree of results," you will be able to see all of the names of the proteins related to your amino acid sequence, which is labeled "unnamed protein product" and highlighted in yellow.
- The amino acid sequence you submitted is labeled "unnamed protein product" and is highlighted yellow. Trace this branch to the left, toward the trunk of the tree, and look at how it connects to the other branches. The less branch and line distance between two proteins, the more closely related they are. For example, two proteins that are right next to each other on the same vertical branch are more closely related than two proteins that are separated by a horizontal branch, or multiple horizontal branches. The more branches/lines you have to trace to reach a node that two proteins have in common on the left side, or the more branches you have to trace to travel between the two proteins, the more distantly related the proteins are. Besides the Niemann-Pick C1 protein, which proteins are most similar to your sequence? Which proteins are the most unrelated?
- Find the two proteins that are the most closely related to your sequence, besides NPC1. Ignore whether they are "isoforms," and only look at the name before the word "isoform." For example, "protein patched homolog 1 isoform S [Homo sapiens]" is the protein called patched homolog 1, and all the isoforms of this protein are just the same protein for the purposes of this science project.
- Write these two protein names in your lab notebook.
- Just how similar are these two proteins to the NPC1 protein? To find out, go back to the NCBI BLAST results page, shown in Figure 5, and locate the names of your two proteins in the list shown in the "Description" tab. In the results table, look what their "Query coverage," "Max Score" and "Precent Identity" is.
- To see exactly how the amino acid sequences match each other, unselect all results in the "Description" tab by clicking on the "Select all" box in the top left corner of the BLAST results table. Then select the two proteins you wrote in your lab notebook and click on the "Alignment" tab. In this section, the NPC1 amino acid sequence you searched for is shown as the "Query" and the protein it is being compared to is the "Sbjct." Amino acids that the two sequences have in common are connected with a vertical line, while there is no line between amino acids that are different in each sequence. Dashes in either sequence indicate a gap in the match. Note the "Identities" value, which is the percent of amino acids/letters that are the same in the query and the subject sequence. If the % identity between two sequences is 97%, then these two sequences differ by 3% in their sequence.
- In the "Description" tab, check the "select all" box again to select all BAST results. Then go to the "Graphic Summary" tab. At the top of the image that says "Distribution of the top ... BLAST Hits on ... subject sequences," the query sequence is shown as a turquoise rectangle. Below the query sequence, the other amino acid sequences that matched it are shown as colored rectangles. The color key shows the degree of similarity for the sequences. Click on the sequence matches to learn more about them.
- Where do the "Patched" proteins match your sequence? Researchers do not know where the drugs bind the NPC1 protein, but where could the drugs bind and probably not also bind the Patched proteins?
- Overall, how likely do you think it is that a drug that binds the NPC1 protein would also bind the Patched proteins?
- You have read about the NPC1 protein, but what do the two similar proteins you picked in step 3h do? Go back to the NCBI Gene website, which you explored in step 1, and instead of searching for NPC1, search for each of these two similar proteins (enter their names instead of their abbreviations, for example "patched homolog 1"). Click on the top search result that is a human gene.
- Read about the genes in the "Summary" section. What do they have in common with NPC1, and how are they different? Based on their function, how damaging do you think it might be if a drug blocked the proteins encoded by these genes? You can also explore the protein page for this gene to find more information.
Investigating Complicated Interactions
Next, you will investigate how even if a drug only binds its target protein, it may still disrupt delicate biological processes. To do this we will look at the signaling pathways (biochemical pathways) that NPC1 and related proteins are involved in. These intricate pathways may be disrupted if these proteins cannot function.
- You can learn about the signaling pathways that NPC1 and related proteins are involved in by using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Database.
- In the search box under "Enter keywords," enter NPC1 and click "Go."
- Click on the "Thumbnail Image" for each pathway result.
- Look at the pathways for NPC1. NPC1 should be in red on the pathway diagrams, but if you cannot locate a particular protein on a pathway diagram, search for its name in the search box at the top of the page.
- Based on the pathway diagrams, how important does NPC1 seem? What cellular processes is it involved in, and is it involved in many or just a few? What downstream events (events that NPC1 leads to) would be affected if NPC1 could not function?
- To learn more about a protein in the pathway, click on its name (inside a box).
- Repeat step 1 using the two proteins you identified in the section titled "Identifying Non-Target Proteins," in step 3h.
- To find out more about what proteins NPC1 and potential non-target proteins interact with, go back to the NCBI Gene website for these proteins.
- As a reminder, in the section titled "Identifying Non-Target Proteins," you already looked at the NCBI Gene website for NPC1 (step 1) and for the two non-target proteins (step 4).
- Once on their NCBI Gene pages, for each gene of interest click on the "Interactions" link, in the "Table of contents" sidebar on the right-hand side of the page.
- The column labeled "Other Gene" lists the names of proteins that interact with the protein of interest. Click on an interacting protein's name and read its "Summary."
- After looking at the functions of the interacting proteins, what other biological processes do you think may be disrupted if NPC1 and/or the two non-target proteins are disrupted?
What Parts of the Body Might the Drugs Affect?
You now probably have a good idea of what biological processes may be disrupted by the drugs targeting NPC1, but what areas of the body may be particularly damaged, and what kinds of people should be most careful to avoid drugs like these? To answer this, we will look at gene expression data of NPC1 and other genes that encode for proteins that may be disrupted by the drug.
- You can learn about the expression of NPC1 and other genes using the NCBI Gene Database again.
- On the NCBI Gene page for the NPC1 gene, scroll down to the "Expression" section or click the "Expression" link in the "Table of contents" on the right side of the page.
- The bar graph in the "Expression" section shows gene expression data for the NPC1 gene in different cell types or tissue samples (Figure 7). On the x-axis, there are cell types or tissue samples listed. For every cell type, the levels of gene expression are shown on the y-axis.
- Which cell types express the highest amounts of NPC1, and which express the lowest? Are there any cell types that do not express it at all?
- If a person took a drug that binds NPC1, what tissues and organs may be most affected? What kinds of people should most avoid taking the drug?
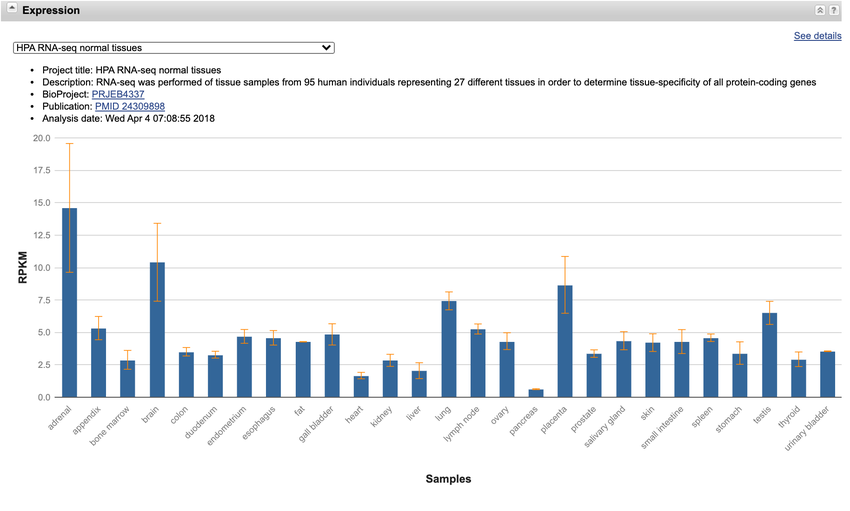 Image Credit: National Center for Biotechnology Information (NCBI)
Image Credit: National Center for Biotechnology Information (NCBI)The 'Expression' section of the NPC1 gene page on the NCBI website has a chart on the levels of gene expression for different cell types or tissue samples. This chart is near the center of the page and shows cells that express a high amount of NPC1.
Figure 7. The chart in the "Expression" section of the NPC1 gene shows the expression levels of NPC1 in different tissue samples or cell types.
- Repeat step 1 using the two proteins you identified in the section titled "Identifying Non-Target Proteins," in step 3h.
Ask an Expert
Global Connections
The United Nations Sustainable Development Goals (UNSDGs) are a blueprint to achieve a better and more sustainable future for all.
Variations
- Drugs can be made to target specific tissues. Search online to find out what tissues the Ebola virus infects the most. A good place to start would be to read the paper titled "Ebola Virus Pathogenesis: Implications for Vaccines and Therapies" that is referenced in the Bibliography above. If anti-viral drugs could be made to only target the infected tissues, would this still be very damaging to the patient, based on the expression data you found in the Experimental Procedure section titled "What Parts of the Body Might the Drugs Affect?"
- Defects in the NPC1 gene are actually associated with a genetic disease. To read more about it, visit the NPC1 Gene webpage. How does this agree, or disagree, with what you have learned about the function of NPC1? Do you think taking Ebola virus drugg could cause similar symptoms? Why or why not? Make sure to tie your reasons to the data you found about the normal and disease functions of NPC1.
- You can investigate the 3D structure of the NPC1 protein using chemistry modeling programs online. Follow the steps below to look up the NPC1 protein and see how it binds cholesterol. Although it is not known where exactly the Ebola drugs bind the NPC1 protein, by looking at the 3D structure of NPC1 and seeing where it binds cholesterol, where do you think a drug could bind the NPC1 protein and still allow the protein to carry out its important role in transporting cholesterol? What areas could the drugs bind that would be the most damaging to the normal function of NPC1?
- First go to the RCSB Protein Data Bank (PDB).
- In the search box at the top, search for "NPC1."
- Look at the different results for NPC1. Many of these results are the 3D structure of NPC1 bound to cholesterol molecules. Click on a result.
- Underneath the protein image on the left side, next to 3D View, click on "Structure" or "Ligand Interaction."
- Here you will see a 3D model of NPC1, possibly interacting with cholesterol. You can rotate the image using left click and drag. You can zoom by scrolling up and down.
- If shown, where is cholesterol binding NPC1?
- Hover your mouse over different parts of the protein to see which amino acid that part of the protein is made of. This is displayed in the bottom right corner of the window.
- For example, "ILE 1210" means that you are hovering over an isoleucine amino acid that is at position 1210 in the protein.
- Can you see which amino acids are interacting with cholesterol the most, and can you determine what kind of bonds are being formed based on your knowledge of the structure of NPC1?
Careers
If you like this project, you might enjoy exploring these related careers:
Related Links
- Science Fair Project Guide
- Other Ideas Like This
- Medical Biotechnology Project Ideas
- Genetics & Genomics Project Ideas
- Big Data Project Ideas
- Pandemics – COVID-19 Project Ideas
- My Favorites
- Genetics Home Reference Tutorial
- NCBI Gene & SNP Tutorial
- PharmGKB Pharmacogenomics Knowledge Base Tutorial