Abstract
Remember going to the doctor and getting vaccine shots? It is no fun getting poked with a needle, but fortunately, a vaccine helps our immune system to develop protection against a serious illness for years to come. But what about the flu vaccine? How come there is a new one every year? This science fair project will show you why.Summary
Sponsor: The Molecular Sciences Institute, Berkeley, CA
Editor: Ken Hess, Science Buddies

Objective
Use free Internet-based computer tools to analyze and estimate the effectiveness of different flu vaccines.Introduction
Influenza, commonly known as the flu, is caused by a virus that attacks the upper respiratory tract (i.e., the nose, the throat and the lungs). Cold and dry weather allows the virus to survive longer outside the body than in warm weather. Therefore, in temperate regions like North America, when we are planning to enjoy Halloween, Thanksgiving, or Christmas, it is also the time when we or our family members have a higher chance of getting the flu.
There are three types of influenza virus: A, B and C. Type A can infect humans, other mammals and birds and can spread fast and affect many people. Types B and C affect only humans and type C causes only a mild infection. Influenza type A viruses are sub-typed into two categories based on proteins, specifically the proteins hemagglutinin and neuraminidase, on the surface of the virus. The virus uses the hemagglutinin protein (often abbreviated "H" or "HA") to latch on to the host's cell and uses the neuramidase protein (often abbreviated "N" or "NA") to spread the infection. Types A and B viruses continually evolve genetically, with changes being made to the amino acid sequence of the H and N proteins. Since hosts recognize the H and N surface proteins to identify and attack the virus, by changing these proteins a little bit the virus prevents the hosts from enjoying any prolonged protection against the virus.
The human immune system has various ways of responding to an infection such as the flu. Our immune system recognizes pathogens like the flu virus and then attacks and destroys them. In a normally functioning immune system, once a pathogen is found, a subset of white blood cells (called B-cells) make antibodies to the pathogen. An antibody is a y-shaped protein and much smaller than even a cell or most pathogens. It both tags a pathogen as "foreign intruder" and helps to destroy the pathogen. It is also important to know that antibodies are usually unique, meaning that flu antibodies cannot grab onto a strep throat caused by bacteria, nor can antibodies to the bacteria identify and destroy a flu virus. This means antibodies are highly specific and bind tightly to only one type of structure on the surface of a cell or pathogen. This antibody binding site is called the antigen. A diagram of antibodies binding onto a pathogen is shown in Figure 1 below. Notice that multiple copies of an antibody will often grab onto a single pathogen. Our bodies produce antibodies that are highly specific for the infectious agent as part of our humoral immune response. The antibodies help stop the infection from spreading further and help to eliminate the pathogen from the body.
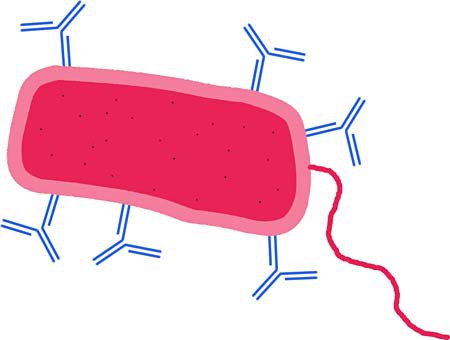
Figure 1. During the immune response, antibodies (shown in blue) bind to a pathogen (a bacterium here, shown in red). Once bound to the pathogen, the antibodies often then get help from white blood cells to destroy the pathogen. Note: These are simplified drawings that are not to scale.
The efficiency of antibodies is based on their ability to specifically bind to their target or antigen. Scientists have made use of this antigen-antibody specificity by developing vaccines that are effective against specific antigens or pathogens such as the flu. Vaccines are a key part of preventative medicine. They prime an individual's acquired immune system so that it has antibodies to recognize and fight off a potential pathogen without ever having to experience the harmful, or even deadly, symptoms of the disease. When a person is vaccinated with the influenza vaccine, it should stimulate a protective immune response, particularly against the viral surface proteins (antigens) in the viral strains used to make the specific vaccine. The influenza vaccine typically contains three virus strains, two are subtypes of type A and one is of type B. Type C is not included in the vaccine because it only causes a mild illness and does not lead to epidemics. To make the influenza vaccine, gene fragments that encode the H and N viral surface proteins (antigens) are used from each strain. Once these antigens enter our body via a flu shot, our immune system starts to produce specific antibodies against all the antigens present in the vaccine. For the vaccine to give a person good protection against the virus, the protein sequences for the H and N proteins that are used in the vaccine should closely match the sequences in the strains the person may be exposed to. Every February, the World Health Organization (WHO), based on the analysis of various laboratories across the globe, will decide what influenza virus strains to include in the vaccine for the new year.
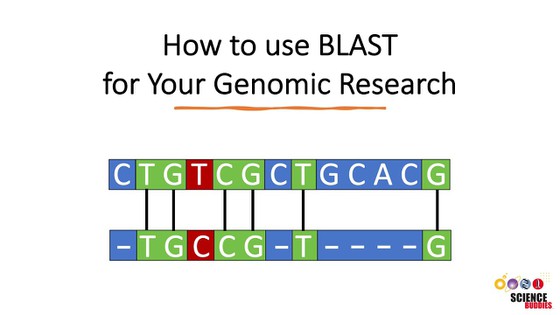
How can scientists check that the protein sequence of the H and N proteins used in the vaccine match the ones in the virus strains they want to protect people against? If you imagine that you can hold the H or N protein with both hands and stretch it out, you will then have a linear protein sequence in your hands. A protein sequence is made up of amino acids. Unlike the English alphabet, which has 26 letters, there are 20 standard amino acids that can be used to "spell" a protein. In English, it is easy to align two words and compare their spellings. Even so, there is often more than one possible alignment, as shown in Figures 2 and 3. In Figure 2, one possible alignment of the words "strawberry" and "blueberry" is shown, where the only matching letter, "r," is highlighted in red.
| s | t | r | a | w | b | e | r | r | y |
| b | l | u | e | b | e | r | r | y | _ |
In Figure 3, another possible alignment of these words is shown, where several matching letters, spelling "berry,"
| s | t | r | a | w | b | e | r | r | y |
| _ | b | l | u | e | b | e | r | r | y |
For the words "strawberry" and "blueberry," the alignment in Figure 3 clearly gives us a greater number of matched letters between these words. Similarly, you can take two protein sequences and compare if their spelling is alike; this is called sequence alignment in bioinformatics.
The alignment example is simple enough that we can do it manually. However, when we want to align two protein sequences, they can be over 100 letters long and consequently it is much more difficult and more time consuming to do it manually. Luckily, bioinformatics comes to the rescue. Bioinformatics is the collection and analysis of large amount of biological data using computers and computational/statistical methods.
A powerful Internet-based bioinformatics tool for aligning sequences is BLAST, which stands for Basic Local Alignment Search Tool. It aligns your query sequence of interest to a collection of sequences stored in the database, or to a specific second sequence you are interested in. It compares the results, telling you which sequences or segments are similar to your query sequence.
All else being equal, we would expect that a strong match between the protein sequences for the H and/or N proteins used in the vaccine virus and the corresponding sequences in the "wild" virus to result in good protection against that virus. On the other hand, a poor match would result in weak protection against the virus. But to create a strong match, the WHO would need to accurately predict which strains people should be vaccinated against for the upcoming flu season. Is the prediction always accurate? How often is there a good match, and how often does the prediction fail and the vaccine does not give good protection against the common strains of the season? In this genetics and genomics science project, you will use BLAST to measure the quality of the match and estimate the effectiveness of a vaccine against different viruses.
Terms and Concepts
- Influenza (or flu)
- Virus
- Influenza virus type
- Surface proteins
- Immune system
- Pathogen
- White blood cell (B-cell)
- Antibody
- Antigen
- Vaccine
- Virus strains
- Epidemic
- Protein sequence
- Amino acids
- Sequence alignment
- Bioinformatics
- BLAST
- Flu notation
Questions
- How are flu viruses named?
- BLAST can be used to align and compare both DNA (nucleotide) sequences and protein (amino acid) sequences. What are some reasons for using a protein alignment instead of a DNA alignment?
- There are many H and N subtypes for influenza A. Why is it that in recent years the annual vaccine has only included influenza A subtypes H1N1 and H3N2? What is happening with the other subtypes? Under what conditions might they be included in the annual vaccine?
- How does a vaccine help prevent the spread of a disease?
Bibliography
To do this science project you will need to use this database.- National Center for Biotechnology Information (NCBI). (August 27, 2012). GenBank Overview. NCBI GenBank, U.S. National Library of Medicine. Retrieved January 18, 2013.
- Centers for Disease Control and Prevention (CDC). (2016, May 4). Selecting Viruses for the Seasonal Influenza Vaccine. Retrieved April 20, 2018.
- NCBI. (2009). National Center for Biotechnology Information. Retrieved August 31, 2009.
- National Center for Biotechnology Information. (2009). Influenza Virus Resources. Retrieved August 31, 2009.
- Centers for Disease Control and Prevention (CDC). (December 28, 2011). Flu Activity & Surveillance. Retrieved September 2, 2009.
Materials and Equipment
- Computer with an Internet connection
- Lab notebook
Experimental Procedure
- First, study the Terms and Concepts in the Background tab. It is especially important that you research and understand flu notation.
- Next, pick an influenza season that you would like to investigate from Table 1. The seasons are listed in the far left column, under "Influenza Season."
- Note that each influenza season spans two years. This is from October of the first year to May of the second year.
- For example, if the influenza season is listed as "2002 - 2003" this means that the data is from October 2002 to May 2003.
- If you want to study more recent flu data for your science project (data that are not included in Table 1), go to the Flu Activity & Surveillance webpage at The U.S. Centers for Disease Control and Prevention (CDC) website.
- Under "Flu Activity & Surveillance" on the left side, click on the link for "Past Weekly Surveillance Reports" and choose the influenza season you want to investigate. Note: Before you choose your influenza season, note that the 2010–2011 influenza report is the last one that contains information on the strains in the next season's (2011–2012) influenza vaccine.
- You will want to investigate a season that has ended so that there is a complete influenza season summary available.
- Click on the "+" sign next to the influenza season you want to investigate. Then click on the link for the season's "Influenza Season Summary" report to open it.
- To find the most common influenza strains subtyped that season, read the section titled "Antigenic Characterization."
- To find information on the strains in that season's influenza vaccine, you will need to go back to the "Past Weekly Surveillance Reports" webpage and select the previous influenza season. For example, if you are investigating the 2010–2011 influenza season, to find information about the 2010–2011 vaccine you will need to look at the 2009–2010 influenza season summary and read the section titled "Composition of the 2010–2011 Influenza Vaccine." Remember that you won't find that information in reports after 2010–2011.
- In your lab notebook, record the years of the influenza season that you chose to investigate.
- Note that each influenza season spans two years. This is from October of the first year to May of the second year.
| Influenza Season | Influenza Type | Most Common Influenza Strains Subtyped That Season | Strains in That Season's Influenza Vaccine |
| 2000–2001 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A/Bayern/07/95 | |||
| A (H3N2) | A/Panama/2007/99 | A/Moscow/10/99 | |
| B | B/Beijing/184/93 | B/Beijing/184/93 | |
| B/Sichuan/379/99 | |||
| 2001–2002 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A (H3N2) | A/Panama/2007/99 | A/Moscow/10/99 | |
| B | B/Yamagata/16/88 | B/Sichuan/379/99 | |
| B/Victoria/2/87 | |||
| 2002–2003 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A (H3N2) | A/Panama/2007/99 | A/Moscow/10/99 | |
| B | B/Hong Kong/330/01 | B/Hong Kong/330/2001 | |
| B/Sichuan/379/99 | |||
| 2003–2004 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A (H3N2) | A/Fujian/411/2002 | A/Moscow/10/99 | |
| A/Panama/2007/99 | |||
| B | B/Hong Kong/330/01 | B/Hong Kong/330/2001 | |
| B/Sichuan/379/99 | |||
| 2004–2005 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A (H3N2) | A/Wyoming/3/2003 | A/Fujian/411/2002 | |
| A/California/7/2004 | |||
| B | B/Yamagata/16/88 | B/Shanghai/361/2002 | |
| B/Victoria/2/87 | |||
| 2005–2006 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A (H3N2) | A/California/7/2004 | A/California/7/2004 | |
| A/Wisconsin/67/2005 | |||
| B | B/Shanghai/361/2002 | B/Shanghai/361/2002 | |
| B/Florida/07/2004 | |||
| B/Ohio/1/2005 | |||
| 2006–2007 | A (H1N1) | A/New Caledonia/20/99 | A/New Caledonia/20/99 |
| A/Solomon Islands/3/2006 | |||
| A (H3N2) | A/Wisconsin/67/2005 | A/Wisconsin/67/2005 | |
| B | B/Ohio/1/2005 | B/Malaysia/2506/2004 | |
| 2007–2008 | A (H1N1) | A/Solomon Islands/3/2006 | A/Solomon Islands/3/2006 |
| A/Brisbane/59/2007 | |||
| A (H3N2) | A/Wisconsin/67/2005 | A/Wisconsin/67/2005 | |
| A/Brisbane/10/2007 | |||
| B | B/Yamagata/16/88 | B/Malaysia/2506/2004 | |
| B/Florida/4/2006 | |||
| B/Ohio/1/2005 | |||
| 2008–2009 | A (H1N1) | A/Brisbane/59/2007 | A/Brisbane/59/2007 |
| A (H3N2) | A/Brisbane/10/2007 | A/Brisbane/10/2007 | |
| B | B/Yamagata/16/88 | B/Florida/4/2006 | |
| B/Victoria/2/87 | |||
| 2009–2010 | A (H1N1) | A/Brisbane/59/2007 | A/Brisbane/59/2007 |
| A/California/07/2009 | |||
| A (H3N2) | A/Brisbane/10/2007 | A/Brisbane/10/2007 | |
| B | B/Brisbane/60/2008 | B/Brisbane/60/2008 | |
| 2010–2011 | A (H1N1) | A/California/07/2009 | A/California/07/2009 |
| A (H3N2) | A/Perth/16/2009 | A/Perth/16/2009 | |
| B | B/Brisbane/60/2008 | B/Brisbane/60/2008 |
- Take a moment to look at the different strains listed in Table 1 for the season you selected. Look at both the strains that were the most common ones subtyped that season as well as the strains that were in that season's vaccine.
- The influenza type of each specific virus strain is listed in the column labeled "Influenza Type," on the same row as the virus strain's name.
- For example, in the 2010–2011 season, the A/California/07/2009 strain is listed as a type A virus, specifically an H1N1 virus. The "H" and "N" refer to the type of hemagglutinin and neuraminidase surface proteins on the virus.
- You may notice that in some seasons there were multiple common strains of the same type (or subtype) of influenza virus. For example, in the 2009–2010 season, there were two common types of Type A (H1N1) strains, specifically A/Brisbane/59/2007 and A/California/07/2009.
- Note how the strains are written in flu notation. What do the notations tell you about the strains?
- For example, in the 2010–2011 season, the strain "A/California/07/2009" is listed. The notation means that this viral strain is a type A virus and it was the 7th influenza virus isolated in 2009 in California.
- The influenza type of each specific virus strain is listed in the column labeled "Influenza Type," on the same row as the virus strain's name.
- In your lab notebook, record all of the data listed in Table 1 for your season of interest. To do this you may want to make a small data table similar to Table 1.
- Next you will compare the sequences for the strains that were common that season to the strains that were used in that season's vaccine. This will show you how good of a match the vaccine was to the strains that were prevalent. But before you do this, check the data table in your lab notebook to see if any of the common strains have the same name as the strains in the vaccine. If they are the same, you will not need to compare their sequences since they should be identical.
- To make this easier for you to spot, in Table 1, if a common strain was different than the ones used in the vaccine, the common strain's name has been bolded.
- For example, in Table 1, all of the common strains for the 2010–2011 influenza season are the same as the strains that were used in the vaccine that year (none of the strains are bolded).
- As another example, in the 2009–2011 influenza season, the A/Brisbane/59/2007 strain was both common and included in the vaccine. However, another Type A (H1N1) strain, A/California/07/2009, was also common but was not included in the vaccine that season. (The A/California/07/2009 strain has consequently been bolded there.)
- If, in your data table, any of the strains common that season were the same as the strains used in the vaccine, do not use that common strain in the next steps of the Procedure.
- Before you continue with the next step, it might be helpful to familiarize yourself with the bioinformatic tools and websites that you are going to use to analyze your influenza strains. You can watch the two videos below to learn more about the BLAST tool and the NCBI website and databases.
- Compare the sequences of one of the common virus strains to the same influenza type that was included in the vaccine that season. You will only be analyzing the sequences for the hemagglutinin and neuraminidase proteins. (If you are unsure of why this is, reread the Introduction in the Background tab.) Obtain the sequences for these strains from the NCBI GenBank website.
- On the top of the webpage next to the search bar, select "Protein" from the drop-down menu.
- Type in the name of one of the common influenza strains (e.g., A/California/07/2009) in the search box from your data table. Include the name of the protein you are looking for. For example, "hemagglutinin A/California/07/2009."
- Look for the full-length protein sequence of the hemagglutinin protein. When you find this result, click on it.
- For example, for the A/California/07/2009 strain, the desired result is listed as "hemagglutinin [Influenza A virus (A/California/07/2009(H1N1))]" and on the next line says "566 aa protein," meaning this protein contains 566 amino acids. Do not select a result that says "partial" in its title, as this is not the full protein.
- If there is no data on the hemagglutinin protein for this strain (the protein is not listed in the results), skip this strain. Start step 7 over using a different common strain from your data table.
- You should now be on a webpage for the hemagglutinin protein for the strain. It should look similar to this hemagglutinin example.
- Copy the accession number for the full-length hemagglutinin protein, listed at the top of the webpage. Record this number in your lab notebook. You may want to add to your existing data table so that it can easily include this information.
- For example, for the A/California/07/2009 strain's hemagglutinin protein, the accession number is ACP44189.
- Repeat steps 7a to 7e but this time in step 7b type in the name of the vaccine strain that is the same type of influenza as the common strain you just searched for.
- For example, if you are investigating the 2009–2010 influenza season and just searched for the A/California/07/2009 strain, you will now want to search for the A/Brisbane/59/2007 strain (the Type A, H1N1, strain used in the vaccine that season).
- Do not forget to record the accession number for hemagglutinin protein for this strain in your lab notebook.
- On the webpage for the hemagglutinin protein of the vaccine strain, click on the "Run BLAST" link (column on right side of webpage, at the top).
- In the "Enter Query Sequence" box, click the box next to "Align two or more sequences." The BLAST alignment input page should look as in Figure 3.
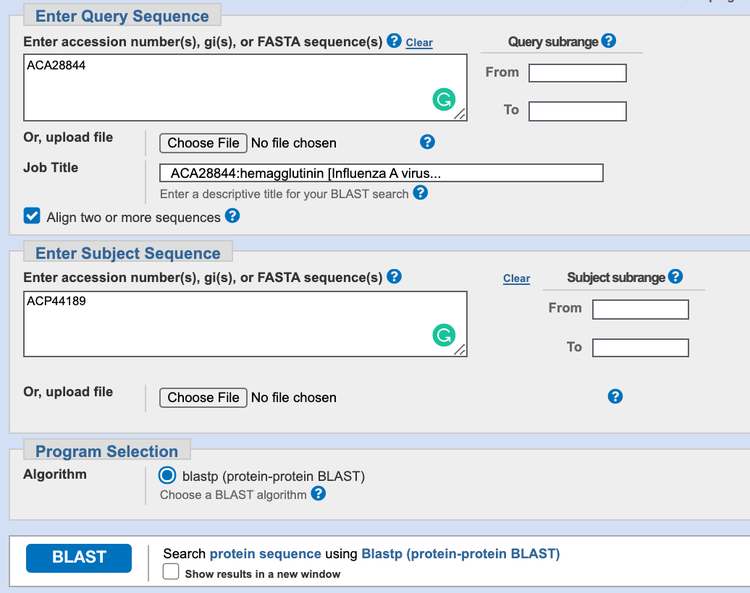 Image Credit: NCBI
Image Credit: NCBI
Figure 3. BLAST two sequence alignment input page. Your query page should look similar to this one after you have filled it in with the two sequences.
- The accession number for the hemagglutinin protein of the vaccine strain should already be filled in in the "Enter Query Sequence" box. In the "Enter Subject Sequence" box below that, enter the accession number for the hemagglutinin protein of the common influenza strain (Figure 3).
- The database should be set automatically to use the algorithm "blastp."
- Next to the BLAST button at the bottom, check the box "Show results in a new window" and then click the BLAST button. You may need to wait a few seconds for your results to appear.
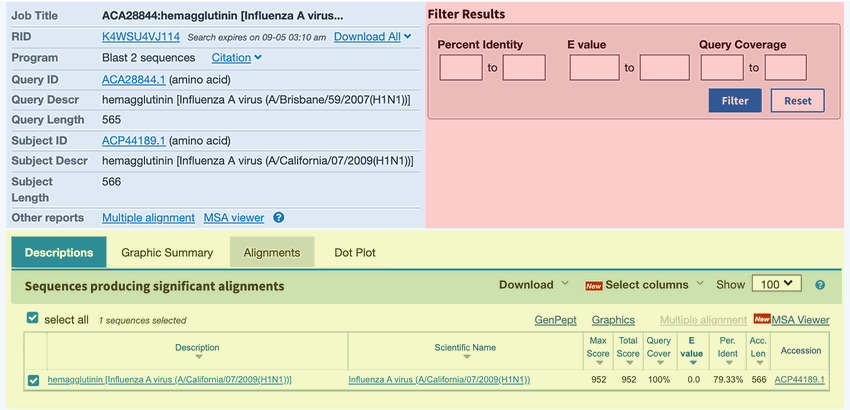 Image Credit: NCBI
Image Credit: NCBI
Figure 4. Snapshot of the BLAST outout page for the two sequence alignment search.
- Look at your BLAST result page (Figure 4). On the top left of the BLAST results page you will find the summary section (blue in Figure 4), which provides information on different aspects of your search. On the top right there is a box that allows you to filter your results based on certain criteria (red in Figure 4). Below the top section, the BLAST results are shown (yellow in Figure 4). There are four different tabs called "Description," Graphic Summary," "Alignments," and "Taxonomy." Each tab presents the search results in a different way.
- The "Description" tab contains a summary table of hits found by BLAST and is the default tab shown. For this science project, the list will only show the subject sequence that you put into your alignment search.
- The "Graphic Summary" tab shows a color key of the alignments. The color key shows the degree of similarity for the sequences.
- The "Alignment" section contains the detailed pairwise alignment between the query and subject sequence.
- The "Taxonomy" section provides details of the taxonomic distribution of matches BLAST found.
- Click on the "Alignment" tab to look at the alignment of your two sequences (Figure 5).
- The top row of letters (labeled "Query") corresponds to the sequence you pasted into the "Query Sequence" box. The bottom row of letters (labeled "Sbjct" or "Subject") corresponds to the sequence you pasted into the "Subject Sequence" box. Where the amino acids/letters at a particular position are the same, a vertical line will connect the two lines of letters. Where there is a mismatch, the letters will NOT be connected by a vertical line.
- Note the "Identities" value, which is the percent of amino acids/letters that are the same in the query and the subject sequence. If the % identity between two sequences is 97%, then these two sequences differ by 3% in their nucleotide sequence.
- How similar are the two sequences to each other? In your lab notebook, record the percentage that is identical between the two sequences.
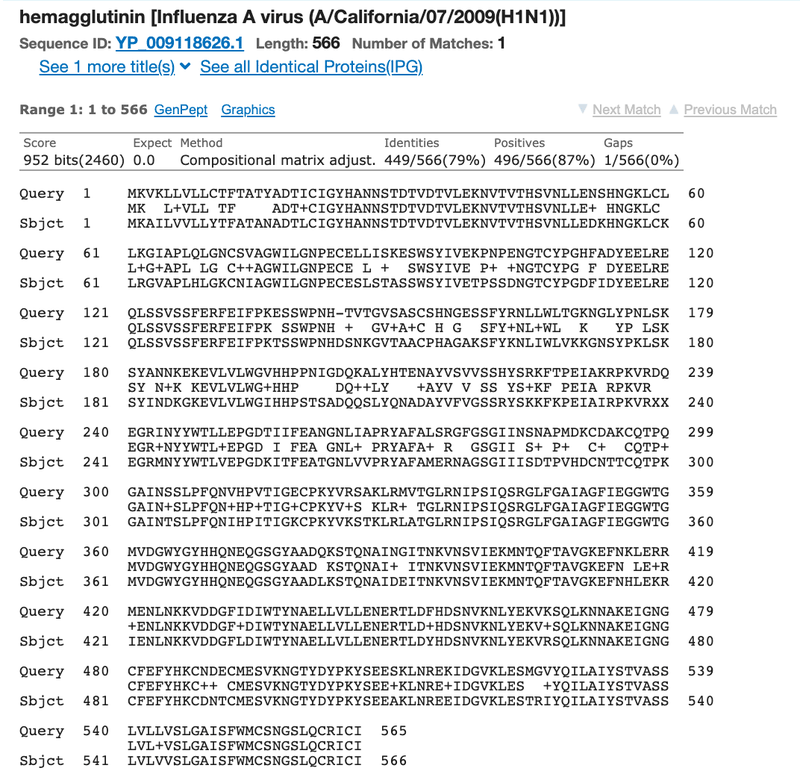 Image Credit: NCBI
Image Credit: NCBI
Figure 5. A detailed view of the two aligned sequences as shown in the "Alignment" tab of the BLAST output page.
- Repeat step 7 until you have compared the hemagglutinin protein of each common strain in your data table with the same type of influenza virus that was used in the vaccination for that season.
- If you want a more advanced challenge, print your BLAST results each time. When you do comparisons between the different strains, see if there is a region of the hemagglutinin protein that tends to be different from the strain used in the vaccination.
- Repeat steps 7 to 8 but this time use the neuraminidase protein sequences for the strains instead of the hemagglutinin protein sequences.
- Repeat steps 2–9 four more times using different influenza seasons (from Table 1) so that you have analyzed a total of five influenza seasons. Do you notice any trends?
- Overall, how similar are the hemagglutinin and neuraminidase protein sequences in the common influenza strains to the same type of influenza virus that was used in the vaccination for that season?
- Based on how similar the sequences are, how well do you think the vaccine protected a vaccinated person from the different strains in a given season?
- How well does it seem that an influenza vaccine from one year will protect a person against the common influenza strains one, two, three, or more years later?
- Do you notice any other trends in your data?
Ask an Expert
Global Connections
The United Nations Sustainable Development Goals (UNSDGs) are a blueprint to achieve a better and more sustainable future for all.
Variations
- Pick an influenza season from Table 1 in the Procedure. If you could travel back in time and redesign the influenza vaccine for the year you pick, which influenza strains would you use for the vaccine? Do you think you could make the vaccine more effective than it was? Based on sequence alignment, if the choice of virus strains you suggest are not available, are there any alternative strains you can use that you think are similar enough to still make an effective vaccine?
- When designing an influenza vaccine, is it important to make sure the vaccine targets certain types of influenza virus more than other types? To figure this out, you can look into how common the different types of influenza virus are each season. The following resource, which is listed in the Bibliography in the Background tab, contains this information. To find the information, follow step 2b in the Procedure. You will need to carefully read through the influenza summary webpage for the season you are interested in. Which types of influenza virus are most common? Does this change from season to season, or does it stay fairly constant?
- Centers for Disease Control and Prevention (CDC). (2009). Flu Activity & Surveillance. Retrieved January 18, 2013, from
Careers
If you like this project, you might enjoy exploring these related careers:
Related Links
- Science Fair Project Guide
- Other Ideas Like This
- Genetics & Genomics Project Ideas
- Medical Biotechnology Project Ideas
- Big Data Project Ideas
- Pandemics – COVID-19 Project Ideas
- My Favorites